Abstract
Schistosoma japonicum is a parasitic flatworm that causes human schistosomiasis, which is a significant cause of morbidity in China and the Philippines. Here we present a draft genomic sequence for the worm. The genome provides a global insight into the molecular architecture and host interaction of this complex metazoan pathogen, revealing that it can exploit host nutrients, neuroendocrine hormones and signalling pathways for growth, development and maturation. Having a complex nervous system and a well-developed sensory system, S. japonicum can accept stimulation of the corresponding ligands as a physiological response to different environments, such as fresh water or the tissues of its intermediate and mammalian hosts. Numerous proteases, including cercarial elastase, are implicated in mammalian skin penetration and haemoglobin degradation. The genomic information will serve as a valuable platform to facilitate development of new interventions for schistosomiasis control.
Similar content being viewed by others
Main
Schistosomiasis is an ancient scourge of mankind, depicted graphically in papyri from Pharaonic Egypt and known from human remains over 2,000 years old from China1,2. Blood-dwelling trematodes (phylum Platyhelminthes) of the genus Schistosoma cause this chronic and debilitating disease, which afflicts more than 200 million people in 76 tropical and subtropical countries. Morbidity is high and schistosomiasis contributes to several hundreds of thousands of deaths annually3,4,5. Three principal species can infect humans: Schistosoma japonicum, Schistosoma mansoni and Schistosoma haematobium. The first of these is prevalent in the Philippines and parts of Indonesia, and is a major disease risk for 66 million people living in southern China2. It remains a major public health concern in China despite over 50 years of concerted campaigns for its control2,6. Approximately one million people in China, and more than 1.7 million bovines and other mammals, are currently infected2. Control measures include community-based praziquantel chemotherapy, health education, improved sanitation, environmental modification and snail control. However, additional approaches, such as the development and deployment of new drugs and anti-schistosome vaccines are urgently needed to meet the prevailing challenges, which include the spectre of praziquantel-resistant parasites7,8.
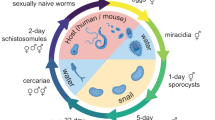
During their complex developmental cycle, schistosomes alternate between a mammalian host and a snail host through the medium of fresh water. After burrowing out of the snail host, free-swimming cercariae penetrate the skin of the mammalian host, travel through the blood to the liver via the lungs, and transform into schistosomula. These mature in the hepatic portal vein, mate and, in the case of S. japonicum, migrate to their final destination in the mesenteric venous plexus. Female worms release thousands of eggs daily, which are discharged in the faeces after a damaging passage through the intestinal wall. If they reach fresh water, eggs hatch to release free-swimming ciliated miracidia, which, guided by light and chemical stimuli, seek amphibious snails of the genus Oncomelania. Within the hemocoel of the snail, miracidia give rise asexually to numbers of sporocysts, in which further asexual propagation produces numerous cercariae.
Eggs deposited by adult female schistosomes embolize in the liver, intestines and other tissue sites and are the key contributors to the pathology and associated morbidity of schistosomiasis. Notably, the highly adapted relationship between schistosomes and their snail intermediate and mammalian definitive hosts appears to involve exploitation by the parasite of host endocrine and immune signals9,10. The evasion strategies that underpin avoidance of the host immune system, allowing schistosomes to survive for years despite strong host immune responses, have long interested investigators intent on development of an efficacious vaccine.
Unlike most other platyhelminths, schistosomes are dioecious. The genome is arrayed on eight pairs of chromosomes, seven pairs of autosomes and one pair of sex chromosomes. Females are the heterogametic sex (ZW); males are homogametic (ZZ)11,12. No other lophotrochozoan13 has yet been sequenced.
Genome features and evolution
General information
The whole-genome shotgun (WGS) sequencing strategy was used to decode the 397-megabase-pair (Mb) sequences, covering most (>90%) of the S. japonicum genome (Supplementary Tables 1 and 2 and Supplementary Fig. 1). A total of 13,469 protein-coding genes were identified, comprising about 4% of the draft S. japonicum genome (Supplementary Figs 2 and 3). Of the protein-coding genes, 6,972 (52%) were mapped to categories established by the Gene Ontology project (Fig. 1a and Supplementary Fig. 4) and an orthologue relationship existed between 2,516 (19%) of them and 1,546 Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology groups (Supplementary Fig. 5). Schistosoma japonicum has a relatively large genome and low gene density in comparison with other invertebrates, including Brugia malayi (Table 1). On the basis of the outbred source of the genomic libraries, high-quality discrepancies found during assembly were used to identify 557,739 single nucleotide polymorphisms (SNPs) (Supplementary Table 3), with an average density of 1.4 SNPs per kilobase pair, and the insertion and deletion (indel) rates were much lower.
a, Proportion of the 6,972 S. japonicum proteins with functional information in different Gene Ontology categories. b, In S. japonicum, vertebrates (H. sapiens, G. gallus and D. rerio), insects (D. melanogaster and A. gambiae), C. elegans and Nematostella vectensis, a total of 7,562 domains were detected. The majority of S. japonicum domains are shared with other taxa, having the fewest unique domains, whereas vertebrates evolved significant numbers of unique protein domains.
Repeat sequences
A total of 657 different repeat families/elements, constituting 159 Mb (40.1%) of the S. japonicum genome were revealed by comparing known repetitive sequences and using the software REPEATSCOUT (version 1.0.3)14 (Fig. 2 and Supplementary Table 4). Among them, 29 kinds of retrotransposon were found, including known Gulliver, SjR1, SjR2 and Sj-pido elements as well as 25 novel elements, together constituting 19.8% of the genome (Supplementary Table 5). Of the 25 novel retrotransposons, 18 are long terminal repeat (LTR) forms, four are non-LTR forms and three are Penelope-like elements—enigmatic retroelements that retain introns15. Each type of retrotransposon was represented by one to 793 intact copies or hundreds to thousands of partial copies. The non-LTR retrotransposons have significantly higher copy numbers, constituting 12.6% of the genome.
Retrons, retrotransposons; SINE, short interspersed nuclear element.
Gene loss/duplication
It was intriguing to observe that schistosomes share more orthologues with the vertebrates (Supplementary Table 6), such as H. sapiens (4,324 pairs), than they do with the ecdysozoans, for example C. elegans (3,292), despite the Ecdysozoa and Lophotrochozoa being phylogenetically adjacent13. Similarly, the cnidarians and vertebrates have been shown to share more orthologous genes with each other than either does with the ecdysozoans16. One possible reason for this is that a higher evolutionary rate in the Ecdysozoa causes an apparently larger orthologue divergence, although the scenario of functional selection of orthologue patterns in the context of parasite–host interplay is also worth consideration.
To test possible consequences of parasitism at the genome level, we investigated gene family and domain variations between schistosomes and other metazoans. It is clear that there was minor variation in total numbers of protein families among S. japonicum (6,322) and the other species, such as C. elegans (6,669), D. melanogaster (5,184) and H. sapiens (6,877) (Supplementary Table 7). However, a major reduction in number, or even the elimination, of protein domains was apparent in the S. japonicum genome, in that the great majority (3,654) of the 3,728 protein domains from the flatworm were shared with other species (Fig. 1b) and can thus be considered ubiquitous among metazoans, whereas 3,834 domains found in at least one of the other species were not detected in schistosomes. Of these 3,834 domains, 1,140 were shared by more than three taxa of vertebrates, insects, a nematode and sea anemones (Supplementary Fig. 6). Notably, domain-loss events seem to be more widespread in S. japonicum than in any other species studied so far, including C. elegans, a model organism well known for rapid evolutionary rates and a high degree of gene loss17. Roughly 1,000 protein domains have been abandoned by S. japonicum, including some involved in basic metabolic pathways and defence, implying that loss of these domains could be, at least partly, a consequence of the adoption of a parasitic way of life.
Against the background of extensive gene/domain loss, the finding of expanded gene families in schistosomes might provide clues to the requirements for a parasitic lifestyle. Among the most expanded gene families in schistosomes (Supplementary Tables 8 and 9), that encoding leishmanolysin (a major surface protease, also called gp63), a member of the metallopeptidase M8 family, has 12 putative family members in S. japonicum, but there is only one in human, fruit fly and nematode (C. elegans), and only three putative counterparts in the free-living flatworm Schmidtea18 (Supplementary Information). In addition to elastase (see later), leishmanolysin-like proteases may contribute to tissue invasion by schistosome cercariae19.
Development and metabolism
Cellular signalling pathways in development
To investigate regulatory networks involved in embryonic development and organogenesis, we undertook comparative genomics analysis of well-characterized signalling pathways, including those for Wnt, notch, hedgehog and transforming growth factor β (TGF-β). Notably, the S. japonicum genome encodes these growth factors, receptors and essential components to regulate many cellular processes during organogenesis and tissue development (Fig. 3 and Supplementary Tables 10 and 11). Schistosoma japonicum also encodes endogenous epidermal growth factor (EGF)-like and fibroblast growth factor (FGF)-like peptides (Fig. 3). The intact downstream cascade composed of the Ras→Raf→mitogen-activated protein kinase (MAPK) and TGF-β→SMAD signalling pathways, including FGF- and EGF-receptors, has components sharing high identity with mammalian orthologues, which implies that schistosomes, in addition to using their own pathways, can exploit host growth factors as developmental signals. Indeed, we have identified an insulin receptor with high sequence similarity with those of mammals20, whereas no insulin growth factor or insulin molecules were found, further supporting the notion that schistosomes exploit key signalling pathways of their hosts for growth and metabolism.
The pathways for growth and development (indicated with different colours), and the neuroactive ligand-receptor interactions in S. japonicum are shown on the left and right, respectively. TACE, tumour-necrosis-factor-α-converting enzyme; ProC, porcupine homologue (Drosophila); NICD, notch intracellular domain; FRP, frizzled-related protein 1; GSK3β, glycogen synthase kinase 3β; TCF, transcription factor 7; ‘p’ within cycle, phosphorylation on the proteins indicated; BMP, bone morphogenetic protein; IGF, insulin-like growth factor; mGlu, metabotropic glutamate; GlyR, glycine receptor; GluR, glutamate receptor; HR, histamine receptor; GABA, γ-aminobutyric acid; DR, dopamine receptor; 5-HT, 5-hydroxytryptamine; HTR, 5-hydroxytryptamine receptor; NPYR, neuropeptide Y receptor; AChR, acetylcholine receptor; RyR, ryanodine receptor; OAR, octopamine receptor; ZIC2, Zic family member 2; CI, cubitus interruptus; suffix ‘R’ denotes receptor.
Metabolic pathways
Analysis of the KEGG pathways assigned to metabolic process (Supplementary Table 12 and Supplementary Figs 7 and 8) indicates that S. japonicum can use carbohydrates as energy/carbon sources. It is unable to de novo synthesize fatty acids, sterols, purines, nine human essential amino acids, arginine or tyrosine (Supplementary Figs 9–11). Loss or degeneracy of fatty acid, sterol and purine synthesis pathways in schistosomes is probably a consequence of the adoption of a parasitic lifestyle; notably, the genes encoding all the key enzymes for both the de novo fatty acid and purine syntheses are complete in the free-living flatworm, Schmidtea mediterranea18 (Supplementary Information). To obtain essential lipid nutrients, the S. japonicum genome indeed encodes many transporters, including apolipoproteins, low-density lipoprotein receptor, scavenger receptor, fatty-acid-binding protein, ATP-binding-cassette transporters and cholesterol esterase (Supplementary Table 13), to exploit fatty acids and cholesterol from host blood and plasma.
Nervous system and neuroendocrine system
Platyhelminths possess a central nervous system with a variety of sensory structures that can transduce a wide range of stimuli, and use a neuroendocrine system to regulate growth, metabolism and homeostasis.
Neurotransmitters and receptors
We characterized a number of receptors and transporters of neurotransmitters (Supplementary Table 14) that may be required, for example, by miracidia and cercariae to navigate through water to locate new hosts and for the schistosomula and adult flukes to establish and reproduce within the human vasculature. In addition to known neurotransmitters and receptors, we have identified a receptor for octopamine (Supplementary Table 14) and two key enzymes for synthesis of octopamine (Supplementary Fig. 12).
The nervous systems of flatworms can be considered to be predominantly peptidergic21. We found additional putative neuropeptide receptors for opioids, galanin and melatonin. Thus, it appears that schistosomes can accept stimulation of the corresponding ligands as a physiological response to different environments, such as fresh water or the tissues of their snail and mammalian hosts. There are genes encoding receptors predicted to accept gastrointestinal neuropeptide hormone signals including cholecystokinin, secretin, gastric inhibitory polypeptide and xenin, all of which are involved in functions promoting the release of alimentary tract fluids containing digestive enzymes.
We also identified receptors for urotensin, angiotensin II and neuromedin (types U and B), which have an important role in physiological regulation of the cardiovascular system, the hypothalamus and other vertebrate organs. Although schistosomes do not have these organs, these components could have other effects on the cells or tissues of the blood fluke, such as the regulation of cell growth or in tissue remodelling. In addition, we found receptors for hypocretin (orexin), leptin and hypothalamic neuropeptides. Together, these features suggest that schistosomes have many advanced physiological features regarded as more characteristic of higher metazoans.
Unexpectedly, a myokinin-like receptor was also observed (Supplementary Table 14). Myokinins are invertebrate neuropeptides with myotropic and diuretic activities for which a receptor, called lymnokinin receptor, was first identified in the tick Boophilus microplus22. The discovery of such a receptor in schistosomes supports the notion that they might synthesize myokinin because their vertebrate hosts do not produce this neuropeptide. Additional examples of receptors found for other invertebrate neuropeptides included FMRFamide and myosuppressin23,24, both belonging to the FMRFamide-like peptide superfamily.
Complex sensory system
Schistosomes have a variety of sensory structures using which they, during their different life stages, presumably respond to a myriad of environmental stimuli. Free-living cercariae and miracidia can sense light, mechanical stimuli and temperature25, facilitating the finding of hosts, whereas the parasitic adult worms are able to respond to changes in levels of chemicals and nutrients. Using a top-down Gene-Ontology-based strategy to facilitate the gene annotation (Supplementary Fig. 13), we identified 71 genes encoding receptors, membrane channels, enzymes and other components, such as rhodopsins/opsins26, phosrestins/arrestins27, transducins, cyclic nucleotide-gated channel, rhodopsin kinase and guanylate cyclase 2D (Supplementary Table 15). Both S. japonicum and S. mansoni have only two members of the rhodopsin family, unlike Drosophila, which possesses 13 members, and zebrafish, which has at least seven (Supplementary Fig. 14a). Phylogenetic analysis indicated that there are at least four schistosome transducins, each of which could represent a divergent subtype of transducin superfamily across chordates, echinoderms, molluscs and arthropods (Supplementary Fig. 14b), and could therefore mediate distinct responses of sensors to signals.
The genome sequence analysis also revealed an array of genes encoding sensory proteins that could interact with chemical ligands and other stimuli. These included guanine-nucleotide-binding protein, potassium-voltage-gated-channel protein Shaker, the glutamate receptor for umami taste and protein Prospero28 (Supplementary Table 16 and Supplementary Fig. 15a). Notably, the genome encodes most components of four of the five human gustatory sensation pathways: the salty, sour, sweet and umami tastes. We also found several potential sensors for sound perception, a common characteristic of vertebrates and arthropods29, in the genome (Supplementary Table 17).
We discovered an apparently intact olfaction pathway, including cyclic nucleotide-gated olfactory channel, guanine-nucleotide-binding protein and adenylyl cyclase type 3 (Supplementary Table 18 and Supplementary Fig. 15b). Moreover, mechanosensory perception mediated by mechanically gated ion channels represents the basis for the sensing of touch, balance, temperature and sound, and contributes essentially to the development and homeostasis of all Eumetazoa30 (Supplementary Table 19). Putative sensory components for equilibrium/balance, mechanical stimulation, pain and temperature (Supplementary Tables 20 and 21) were also found in the S. japonicum genome, including two proteins that have similarities with the well-known mechanosensory protein, transient receptor potential cation channel31, and several receptors such as metabotropic glutamate receptor 3, which participate in the sensory perception of pain, light and taste.
Neuroendocrine system
Schistosomes have receptors that apparently evolved to accept endogenous hormones as well as those of the parasitized mammalian host20,32. By surveying hormones and receptors related to the classical neuroendocrine axis in the genomic sequence of S. japonicum, we found (Fig. 4) putative receptors for hypothalamic hormones such as thyrotropin-releasing hormone (TRH), prolactin-releasing hormone, somatostatin, melanin-concentrating hormone and leptin, as well as transmembrane proteins that have some similarities with receptors for gonadotropin-releasing hormone, corticotropin-releasing hormone and growth-hormone-releasing hormone. Moreover, putative receptors are present that show weak similarity with those in mammals for the pituitary hormones thyroid-stimulating hormone (TSH), luteinizing hormone, follicle-stimulating hormone, arginine vasopressin and oxytocin.
Structured according to the proposed hypothalamus–pituitary–peripheral-endocrine-glands axis with putative ligands found in S. japonicum coloured in orange and S. japonicum receptors in yellow. CRH, corticotrophin-releasing hormone; GHRH, growth-hormone-releasing hormone; TRH, thyrotropin-releasing hormone; PRH, prolactin-releasing hormone; GnRH, gonadotropin-releasing hormone; TSH, thyroid-stimulating hormone; FSH, follicle-stimulating hormone; LH, luteinizing hormone; suffix ‘R’ denotes receptor.
Although a hypothalamus–pituitary-like organ has not been described in schistosomes, it is possible that some neurons, similar to those in the hypothalamus and pituitary of vertebrates, could fulfil similar functions in terms of modulating the behaviour of S. japonicum through peripheral endocrine tissues and cells. In this regard, it is noteworthy that the genomic information suggests the presence of an integral hypothalamic–pituitary–thyroid axis in S. japonicum. In addition to the superior TRH–TSH receptors, an intact system for synthesis of thyroxine and active triiodothyronine, as well as an inactivation mechanism of these hormones using deiodination, was identified. Nuclear receptors for triiodothyronine and thyroxine were revealed with identity to mammalian orthologues. Hence, S. japonicum may use an endogenous thyroid hormone/receptor signalling pathway for growth and development (Fig. 4 and Supplementary Table 22).
We confirmed that S. japonicum has receptors for steroid hormones such as progestin, progesterone and oestrogen32,33. In addition, it possesses intricate pathways for processing steroid hormones to form other sex hormones. For example, there are putative enzymes present that could convert the female hormones progesterone and pregnenolone to estriol, oestrone, androsterone and testosterone. Hence, schistosomes might use these pathways during their parasitic existence. Schistosoma japonicum also encodes enzymes to catabolize excessive or used steroid hormones such as aldosterone.
With regard to the process of glycolysis for essential energy supply, receptors for adiponectin, an insulin-sensitizing hormone34, and leptin, a suppressor of the secretion of insulin35, are also encoded by the genome of S. japonicum (Supplementary Table 22), providing further support for the notion that the blood fluke modulates its energy metabolism in response to either its own insulin-like hormones or those of its mammalian host.
The schistosomulum renews its tegument during maturation into an adult schistosome under the effects of ecdysone36,37. In concordance, we identified an ecdysone-like receptor and its downstream effector ecdysone-induced protein 78C. In addition, allatostatin, a polypeptide hormone that suppresses the secretion of juvenile hormone, was previously reported to be found throughout the schistosome nervous system38,39. An allatostatin-like receptor sequence that has high similarity with that of the cockroach was also identified (Supplementary Table 22).
Disease pathogenesis
Cercarial elastase and protease superfamily
Schistosome proteases have key roles in invasion40, migration41 and feeding/nutrition42. We identified 314 putative proteases, including metallo-, cysteine, serine, threonine and aspartic proteases, in the S. japonicum genome data set (Fig. 5a and Supplementary Tables 23–27) by searching in the MEROPS database of peptidases. We classified 108 S. japonicum metalloproteases into 21 subtypes, 16 belonging to the aminopeptidases (Supplementary Table 23). Notably, the leucine aminopeptidase of the M17 family was reported as a major egg antigen43,44 and a putative anti-fluke vaccine45. The second largest assemblage comprised the cysteine proteases, of which 102 members were assigned to 17 subtypes (Supplementary Table 24). Among them, the cathepsins B, C, F and L have pivotal roles in schistosome feeding and nutrition42, as well as in migration through human tissues41. The cysteine proteases cathepsins K and S, as well as the cathepsin A serine protease, have not previously been recognized in schistosomes, and may contribute to catabolism of haemoglobin and other host proteins.
a, The pie chart shows the distribution of the five kinds of protease. b, The genomic structure of S. japonicum cercarial elastase (SjCE). c, A phylogeny of the elastase family in schistosomes using the neighbour-joining method. Bootstrap values are provided above the branches. SmCE, S. mansoni cercarial elastase; ShCE, S. haematobium cercarial elastase; SdCE, Schistosomatium douthitti elastase. d, Immunofluorescence assay showing the presence (white arrow) of SjCE around a schistosomulum following its penetration through mouse skin (panel 2). A naive rabbit serum was used as negative control (panel 4). The location of the cercaria is indicated (white arrow). Panels 1 and 3 show the skin tissue slices under the optical microscope.
Among the 65 serine proteases (Supplementary Table 25), we discovered a S. japonicum cercarial elastase (SjCE), an enzyme that in S. mansoni is vital in the penetration by cercariae of mammalian skin to initiate infection40,46. The elastase locus predicted from the S. japonicum genome spans three exons and two introns, similar to the known S. mansoni elastases47 (Fig. 5b); however, unlike for S. mansoni, only a single elastase was identified in S. japonicum. Phylogenetic analysis of available schistosome elastases (Supplementary Table 28) suggested that the elastase genes in S. mansoni have expanded through at least two rounds of gene duplication, whereas SjCE is an orthologue of S. mansoni cercarial elastase 2b (Fig. 5c). Moreover, by re-examination of mass spectra data that we collected previously33, we identified a unique peptide (IAFLALSDFDHR) of SjCE in cercariae (Supplementary Fig. 16a). We also confirmed the existence of SjCE gene products in both the sporocyst and cercarial stages of S. japonicum by immunoblot and immunofluorescence assays (Fig. 5d and Supplementary Fig. 16b). In addition, the native protease was recognized by anti-recombinant SjCE antibodies in infected mouse skin, indicating that this cercarial elastase is secreted/released by the parasite during invasion of mammalian skin.
Immune system and inflammatory factors
The immune system of S. japonicum has to face both invading microbial pathogens and the immune statuses of both its molluscan and mammalian hosts. Although adaptive immune molecules such as immunoglobulin are lacking in S. japonicum and a classical Toll-like receptor was not found, putative Toll-interacting protein or proteins containing Toll/interleukin-1 resistance motif or leucine-rich repeats appear to be present (Supplementary Table 29). Therefore, schistosomes, like nematodes, appear to possess a primordial Toll pathway as a first line of defence against microbial infections. The identification of the downstream components of a Toll-related pathway, including putative interleukin-1-receptor-associated kinases, toll-like receptor adaptors, TNF-receptor-associated factor 6 (TRAF6), inhibitor of nuclear factor κB kinase subunit epsilon (IKK-ε) and p38 MAPK, further support the view that this primitive innate immune system could be crucial for the worm (Supplementary Table 30).
On the other hand, factors and metabolites in S. japonicum that could contribute to stimulation and regulation of mammalian immunity were discovered. It is well accepted that glycans and lipids synthesized by adult schistosomes or eggs may regulate secondary signals through corresponding receptors on effector cells and accessory cells of the mammalian host, thus compromising host immunological defences targeting the parasite. We therefore searched for enzymes involved in the metabolism of various glycans or lipid antigens by interrogating this worm genome. It turned out that, with the rare exception of enzymes such as α1,3-mannosyltransferase, a complete set of enzymatic machinery for biosynthesis and modification of glycans and lipids exists (Supplementary Table 31).
In addition, prostaglandins, which are well-known mediators of inflammation, can be synthesized by S. japonicum as a result of arachidonic-acid metabolism. It is feasible that S. japonicum synthesizes arachidonate by using lecithin, converting the arachidonate into leukotriene A4 using arachidonate 5-lipoxygenase, followed by the conversion of unstable leukotriene A4 into the active chemical leukotriene B4 through leukotriene A4 hydrolase. The S. japonicum genome also encodes putative receptors for leukotriene B4, cysteinyl leukotriene and prostaglandins E2 and F2, suggesting that prostaglandins could have an important role in the physiology of schistosomes and also in the host–parasite interplay. Unexpectedly, S. japonicum possesses proteins paralogous to mammalian autoimmune-disease-related autoantigens; these include 69 kDa islet cell autoantigen (ICA1), islet antigen-2 (PTPRN) and glutamate decarboxylase (GAD), known autoantigens related to type-I diabetes in β-cells, which raises the possibility that these autoantigen-mimicking molecules could induce chemokine-receptor-mediated cell migration and initiate leukocyte migration into inflamed tissue, which ultimately contribute to the granuloma formation that promotes parasite survival.
Concluding remarks
Lophotrochozoa, of which S. japonicum is a member, is a large taxon that includes ∼50% of all animal phyla. Our work provides a model for evaluating the genomic architecture, biology and evolution in this major taxon. Although the genome of S. japonicum has undergone significant protein-domain-loss events, a detailed molecular repertoire exists to permit the pathogen to locate and penetrate hosts, nourish itself and interact with the environment and its host. With the release and analysis of the S. mansoni genome48, a comparative-genomics approach elucidating the similarities and differences between these two closely related parasites will provide more clues regarding these important pathways. Further functional analysis, using approaches such as RNA interference and translational studies are essential to resolve uncertainties in the molecular physiology of schistosomes and to illuminate mechanisms of pathogenesis in schistosomiasis, efforts that may lead to the development of new interventions for its control and eventual elimination.
Methods Summary
We obtained adult worms and eggs of S. japonicum from infected rabbits. The genomic DNA was extracted from ∼1,000 mixed, outbred adult male and female S. japonicum, perfused from rabbits infected with cercariae released by naturally infected snails. Genomic libraries, including bacterial artificial chromosome (BAC), fosmid and plasmid libraries, were constructed. We performed WGS sequencing on capillary sequencers, and then used a modified PHUSION (version 2.1c) package to assemble the reads. Protein-encoding genes were predicted using EXONHUNTER (version 2.0)49. We used a stepwise method to predict the gene functions. The metabolic and regulatory pathway of S. japonicum was reconstructed with reference to the KEGG pathway database. Proteins were first clustered using a Markov cluster algorithm and then merged according to protein-domain information to establish protein-family clusters. We used immunoblot and immunofluorescence assays to detect cercarial elastase.
Online Methods
Schistosoma japonicum genomic and full-length cDNA library construction
Genomic DNA was extracted from ∼1,000 mixed, outbred adult male and female S. japonicum, perfused from rabbits infected with cercariae released by naturally infected snails collected from an endemic focus in Anhui Province, as described20. Four genomic libraries with different insert sizes were constructed, one of bacterial artificial chromosomes (inserts, 80–120 kb), one of fosmids (36–42 kb) and two of plasmids (6–10 kb and 1.6–4 kb) (Supplementary Table 1). Total RNAs from S. japonicum adults and eggs were isolated using Trizol (Invitrogen), after which mRNA was purified using the Poly(A) Purist mRNA Purification Kit (Ambion). Two full-length cDNA libraries, from adults and eggs were constructed using a modified biotinylated CAP-trapper approach52,53.
WGS sequencing and assembly
After the clone ends of four discrete genomic libraries were sequenced by capillary DNA sequencers ABI3700 (Applied Biosystems) and MegaBACE 1000 or MegaBACE 4000 (General Electric), PHRED (version 0.020425.c)54,55 was used for base calling. All reads were qualified by removing clone vector and bacterial host sequences, as well as the host rabbit (Oryctolagus cuniculus) DNA sequences (http://www.ensembl.org/Oryctolagus_cuniculus/index.html). A modified PHUSION (version 2.1c) package56 was used for assembly.
Repeat and retrotransposon identification
A repetitive sequence library of S. japonicum was generated by the method of consensus seed extending using REPEATSCOUT (version 1.0.3)14, with the k-mer size of 16. Tandem repeats in the genome were identified using TANDEM REPEATS FINDER (version 4.00)57 and categorized using the tandem repeats analysis program TRAP (version 1.0)58. Microsatellites, minisatellites and satellites are classically defined as repeat units of 1–6 bp, 11–100 bp and more than 100 bp, respectively. Polyprotein and reverse transcriptase from GenBank were used as queries to search genome sequences of S. japonicum using tBLASTN (e-value ≤ 10-10). The best hit sequences were then used to query the genome, and those yielding multiple hits in the genome were categorized as candidate retrotransposons. All candidate retrotransposons were assembled to establish complete CDSs encoding polyprotein or reverse transcriptase. Once the complete CDS was determined, sequences upstream and downstream of this CDS in the genome were analysed to identify LTRs which flank the left and right termini of LTR retrotransposons and retroviruses.
Prediction and integration of protein-coding genes
Protein encoding genes were predicted using EXONHUNTER (version 2.0)49. The prediction program combined ab initio gene prediction with supporting evidence from S. japonicum and S. mansoni expressed sequence tags, S. japonicum pair-end ditags, the Swiss-Prot protein database59 and the Pfam protein-domain database (version 22.0)60. Because there were few training sets available for S. japonicum or for any other closely related species, we developed an iterative method that started from the distantly related species C. elegans, and progressively improved parameters of the gene finder on the basis of well-supported predicted gene fragments. The predicted genes were merged with putative expressed sequence tags and full-length cDNA-derived CDSs (proteins), yielding an integrated protein-coding gene set for further functional analysis. These genes were classified into categories established by the Gene Ontology project through the encoding proteins or domains matched to the Gene Ontology index provided by UniProt61 and InterPro62 (iprscan_DATA_17.0 and iprscan_PTHR_DATA_14.0).
Genome variation analysis
The PHUSION assembler56 does not provide alignment information of reads to its contig consensus, so BLASTN was used to relocate reads to contig consensus, with overall identity of over 95%, and to provide alignment information. We established a locally developed SNP pipeline based on neighbourhood quality standard, with the following rules: for each candidate SNP on shotgun reads, the 5-bp flanking sequences should be the same as the contig consensus, the base quality on the SNP site should be no less than 23 and the base quality of the flanking 5 bp should be not less than 15 (refs 63, 64).
Pathway mapping
The metabolic and regulatory pathway of S. japonicum was reconstructed on the basis of the KEGG pathway database65. The KEGG orthology identifier was used as a linkage between genes and pathways. The assignment of S. japonicum genes to KEGG orthologues was implemented with a modified bidirectional-best-BLAST-hits method, which was adjusted using phylogenetic information. The pathway mapping results for the S. japonicum genome are available at http://chgc.sh.cn/japonicum.
Gene-family analysis
Proteins of S. japonicum, C. elegans, D. melanogaster, A. gambiae, D. rerio, G. gallus, H. sapiens and N. vectensis were first clustered using a Markov cluster algorithm66 and then merged according to protein-domain information to establish protein-family clusters. The S. japonicum protein domains were scanned using INTERPROSCAN62. Protein-domain information on other species was sourced from the KEGG database65.
Analysis of S. japonicum proteases
Putative proteases in the S. japonicum data set were identified by comparing S. japonicum cDNA and predicted genes with the MEROPS database67. The results were manually checked and compared with annotations generated by BLAST searches against more comprehensive databases as above. Results with inconsistent annotations from MEROPS and BLAST were removed. For phylogenetic and evolutionary analyses of gene families, deduced amino-acid sequences were aligned using CLUSTAL W (version 1.83)68. Phylogenetic trees were generated using MEGA (version 3.1)69 with the neighbour-joining method and tested with 1,000 bootstrap replicates.
Immunofluorescence assay of S. japonicum cercarial elastase
A mouse anaesthetized with pentobarbital was infected with S. japonicum cercariae. After 10 min, the skin was excised, finely diced, and embedded in OCT fixative. The prepared 7-µm-thick frozen sections were incubated for 30 min in a solution of 20% goat serum in Tris-HCl-buffered saline. The sections were incubated with the rabbit primary antiserum raised against purified recombinant SjCE or normal rabbit serum, followed by a FITC-conjugated second antibody. Fluorescence was visualized using a Leica DM-2500 fluorescence microscope.
Accession codes
Primary accessions
EMBL/GenBank/DDBJ
Data deposits
The sequences of S. japonicum WGS assembly contigs and scaffolds, BACs, full-length complementary DNAs and retrotransposons have been deposited in the European Molecular Biology Laboratory (EMBL) Nucleotide Sequence Database (http://www.ebi.ac.uk/embl/) and the Shanghai Center for Life Science & Biotechnology Information (LSBI; http://lifecenter.sgst.cn/schistosoma/en/schistosomaCnIndexPage.do), and can be freely downloaded. The EMBL accession numbers are CABF01000001–CABF01095265 (contigs) FN330975–FN356022 (scaffolds), FN293020–FN293041 (BACs), FN313573–FN330973 (full-length cDNAs), FN356203–FN356227 (retrotransposons). The LSBI accession numbers are CNUS0000108051–CNUS0000203315 (contigs), CCON0000096785–CCON0000121832 (scaffolds), CNUS0000095394–CNUS0000108050 (predicted genes), CPRT0000000001–CPRT0000012657 (predicted proteins), CNUS0000203316–CNUS0000203337 (BACs), CNUS0000203338–CNUS0000220738 (full-length cDNAs), CNUS0000220739–CNUS0000220763 (retrotransposons). The sequences of S. japonicum integrated protein-coding genes are available on the Chinese National Human Genome Center at Shanghai website (http://www.chgc.sh.cn/japonicum). The BAC library (CHORI-108) is available from the laboratory of P. De Jong at the BACPAC Resources Center, Children’s Hospital Oakland Research Institute, California (http://bacpac.chori.org/library.php?id=168). This paper is distributed under the terms of the Creative Commons Attribution-Non-Commercial-Share Alike licence, and is freely available to all readers at http://www.nature.com/nature.
References
Adamson, P. B. Schistosomiasis in antiquity. Med. Hist. 20, 176–188 (1976)
Zhou, X. N. et al. The public health significance and control of schistosomiasis in China - then and now. Acta Trop. 96, 97–105 (2005)
King, C. H., Dickman, K. & Tisch, D. J. Reassessment of the cost of chronic helmintic infection: a meta-analysis of disability-related outcomes in endemic schistosomiasis. Lancet 365, 1561–1569 (2005)
Steinmann, P., Keiser, J., Bos, R., Tanner, M. & Utzinger, J. Schistosomiasis and water resources development: systematic review, meta-analysis, and estimates of people at risk. Lancet Infect. Dis. 6, 411–425 (2006)
Finkelstein, J. L., Schleinitz, M. D., Carabin, H. & McGarvey, S. T. Decision-model estimation of the age-specific disability weight for Schistosomiasis japonica: a systematic review of the literature. PLoS Negl. Trop. Dis. 2, e158 (2008)
Utzinger, J., Zhou, X. N., Chen, M. G. & Bergquist, R. Conquering schistosomiasis in China: the long march. Acta Trop. 96, 69–96 (2005)
Li, Y. S. et al. Large water management projects and schistosomiasis control, Dongting Lake region, China. Emerg. Infect. Dis. 13, 973–979 (2007)
Bergquist, R., Utzinger, J. & McManus, D. P. Trick or treat: the role of vaccines in integrated schistosomiasis control. PLoS Negl. Trop. Dis. 2, e244 (2008)
Amiri, P. et al. Tumour necrosis factor α restores granulomas and induces parasite egg-laying in schistosome-infected SCID mice. Nature 356, 604–607 (1992)
Davies, S. J. et al. Modulation of blood fluke development in the liver by hepatic CD4+ lymphocytes. Science 294, 1358–1361 (2001)
Hirai, H. et al. Chromosomal differentiation of the Schistosoma japonicum complex. Int. J. Parasitol. 30, 441–452 (2000)
Moné, H. & Boissier, J. Sexual biology of schistosomes. Adv. Parasitol. 57, 89–189 (2004)
Halanych, K. M. The new view of animal phylogeny. Annu. Rev. Ecol. Evol. Syst. 35, 229–256 (2004)
Price, A. L., Jones, N. C. & Pevzner, P. A. De novo identification of repeat families in large genomes. Bioinformatics 21, i351–i358 (2005)
Arkhipova, I. R., Pyatkov, K. I., Meselson, M. & Evgen’ev, M. B. Retroelements containing introns in diverse invertebrate taxa. Nature Genet. 33, 123–124 (2003)
Putnam, N. H. et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317, 86–94 (2007)
Gamulin, V., Muller, I. M. & Muller, W. E. Sponge proteins are more similar to those of Homo sapiens than to Caenorhabditis elegans. Biol. J. Linn. Soc. 71, 821–828 (2000)
Robb, S. M., Ross, E. & Sanchez Alvarado, A. SmedGD: the Schmidtea mediterranea genome database. Nucleic Acids Res. 36, D599–D606 (2008)
Curwen, R. S., Ashton, P. D., Sundaralingam, S. & Wilson, R. A. Identification of novel proteases and immunomodulators in the secretions of schistosome cercariae that facilitate host entry. Mol. Cell. Proteomics 5, 835–844 (2006)
Hu, W. et al. Evolutionary and biomedical implications of a Schistosoma japonicum complementary DNA resource. Nature Genet. 35, 139–147 (2003)
Mousley, A., Maule, A. G., Halton, D. W. & Marks, N. J. Inter-phyla studies on neuropeptides: the potential for broad-spectrum anthelmintic and/or endectocide discovery. Parasitology 131, S143–S167 (2005)
Holmes, S. P., Barhoumi, R., Nachman, R. J. & Pietrantonio, P. V. Functional analysis of a G protein-coupled receptor from the southern cattle tick Boophilus microplus (Acari: Ixodidae) identifies it as the first arthropod myokinin receptor. Insect Mol. Biol. 12, 27–38 (2003)
Egerod, K. et al. Molecular cloning and functional expression of the first two specific insect myosuppressin receptors. Proc. Natl Acad. Sci. USA 100, 9808–9813 (2003)
Scholler, S. et al. Molecular identification of a myosuppressin receptor from the malaria mosquito Anopheles gambiae. Biochem. Biophys. Res. Commun. 327, 29–34 (2005)
Cohen, L. M., Neimark, H. & Eveland, L. K. Schistosoma mansoni: response of cercariae to a thermal gradient. J. Parasitol. 66, 362–364 (1980)
Hoffmann, K. F., Davis, E. M., Fischer, E. R. & Wynn, T. A. The guanine protein coupled receptor rhodopsin is developmentally regulated in the free-living stages of Schistosoma mansoni. Mol. Biochem. Parasitol. 112, 113–123 (2001)
Matsumoto, H. & Yamada, T. Phosrestins I and II: arrestin homologs which undergo differential light-induced phosphorylation in the Drosophila photoreceptor in vivo. Biochem. Biophys. Res. Commun. 177, 1306–1312 (1991)
Grosjean, Y., Lacaille, F., Acebes, A., Clemencet, J. & Ferveur, J. F. Taste, movement, and death: varying effects of new prospero mutants during Drosophila development. J. Neurobiol. 55, 1–13 (2003)
Robert, D. & Gopfert, M. C. Acoustic sensitivity of fly antennae. J. Insect Physiol. 48, 189–196 (2002)
Walker, R. G., Willingham, A. T. & Zuker, C. S. A Drosophila mechanosensory transduction channel. Science 287, 2229–2234 (2000)
Mutai, H. & Heller, S. Vertebrate and invertebrate TRPV-like mechanoreceptors. Cell Calcium 33, 471–478 (2003)
Hu, W., Brindley, P. J., McManus, D. P., Feng, Z. & Han, Z. G. Schistosome transcriptomes: new insights into the parasite and schistosomiasis. Trends Mol. Med. 10, 217–225 (2004)
Liu, F. et al. New perspectives on host-parasite interplay by comparative transcriptomic and proteomic analyses of Schistosoma japonicum. PLoS Pathog. 2, e29 (2006)
Heilbronn, L. K., Smith, S. R. & Ravussin, E. The insulin-sensitizing role of the fat derived hormone adiponectin. Curr. Pharm. Des. 9, 1411–1418 (2003)
Kieffer, T. J., Heller, R. S., Leech, C. A., Holz, G. G. & Habener, J. F. Leptin suppression of insulin secretion by the activation of ATP-sensitive K+ channels in pancreatic beta-cells. Diabetes 46, 1087–1093 (1997)
Foster, J. M., Mercer, J. G. & Rees, H. H. Analysis of ecdysteroids in the trematodes, Schistosoma mansoni and Fasciola hepatica. Trop. Med. Parasitol. 43, 239–244 (1992)
Basch, P. F. Immunocytochemical localization of ecdysteroids in the life history stages of Schistosoma mansoni. Comp. Biochem. Physiol. Comp. Physiol. 83, 199–202 (1986)
Smart, D. et al. Localization of Diploptera punctata allatostatin-like immunoreactivity in helminths: an immunocytochemical study. Parasitology 110, 87–96 (1995)
Smart, D. et al. Peptides related to the Diploptera punctata allatostatins in nonarthropod invertebrates: an immunocytochemical survey. J. Comp. Neurol. 347, 426–432 (1994)
Dvorak, J. et al. Differential use of protease families for invasion by schistosome cercariae. Biochimie 90, 345–358 (2008)
Dvorak, J. et al. Multiple cathepsin B isoforms in schistosomula of Trichobilharzia regenti: identification, characterisation and putative role in migration and nutrition. Int. J. Parasitol. 35, 895–910 (2005)
Koehler, J. W., Morales, M. E., Shelby, B. D. & Brindley, P. J. Aspartic protease activities of schistosomes cleave mammalian hemoglobins in a host-specific manner. Mem. Inst. Oswaldo Cruz 102, 83–85 (2007)
Abouel-Nour, M. F., Lotfy, M., El-Kady, I., El-Shahat, M. & Doughty, B. L. Localization of leucine aminopeptidase in the Schistosoma mansoni eggs and in liver tissue from infected mice. J. Egypt. Soc. Parasitol. 35, 147–156 (2005)
Xu, Y. Z., Shawar, S. M. & Dresden, M. H. Schistosoma mansoni: purification and characterization of a membrane-associated leucine aminopeptidase. Exp. Parasitol. 70, 124–133 (1990)
Hillyer, G. V. Fasciola antigens as vaccines against fascioliasis and schistosomiasis. J. Helminthol. 79, 241–247 (2005)
Newport, G. R. et al. Cloning of the proteinase that facilitates infection by schistosome parasites. J. Biol. Chem. 263, 13179–13184 (1988)
Salter, J. P. et al. Cercarial elastase is encoded by a functionally conserved gene family across multiple species of schistosomes. J. Biol. Chem. 277, 24618–24624 (2002)
Berriman, M. et al. The genome of the blood fluke Schistosoma mansoni. Nature 10.1038/nature08160 (this issue)
Brejova, B. et al. Finding genes in Schistosoma japonicum: annotating novel genomes with help of extrinsic evidence. Nucleic Acids Res. 37, e52 (2009)
Stricklin, S. L., Griffiths-Jones, S. & Eddy, S. R. C. elegans noncoding RNA genes. WormBook 25, 1–7 (2005)
Ghedin, E. et al. Draft genome of the filarial nematode parasite Brugia malayi. Science 317, 1756–1760 (2007)
Seki, M., Carninci, P., Nishiyama, Y., Hayashizaki, Y. & Shinozaki, K. High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J. 15, 707–720 (1998)
Wei, C. L. et al. 5′ long serial analysis of gene expression (LongSAGE) and 3′ LongSAGE for transcriptome characterization and genome annotation. Proc. Natl Acad. Sci. USA 101, 11701–11706 (2004)
Ewing, B., Hillier, L., Wendl, M. C. & Green, P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8, 175–185 (1998)
Ewing, B. & Green, P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8, 186–194 (1998)
Mullikin, J. C. & Ning, Z. The phusion assembler. Genome Res. 13, 81–90 (2003)
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999)
Sobreira, T. J., Durham, A. M. & Gruber, A. TRAP: automated classification, quantification and annotation of tandemly repeated sequences. Bioinformatics 22, 361–362 (2006)
Gasteiger, E. et al. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 (2003)
Finn, R. D. et al. The Pfam protein families database. Nucleic Acids Res. 36, D281–D288 (2008)
UniProt Consortium The universal protein resource (UniProt). Nucleic Acids Res. 36, D190–D195 (2008)
Zdobnov, E. M. & Apweiler, R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17, 847–848 (2001)
Mullikin, J. C. et al. An SNP map of human chromosome 22. Nature 407, 516–520 (2000)
Altshuler, D. et al. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 407, 513–516 (2000)
Kanehisa, M. et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484 (2008)
Enright, A. J., Van Dongen, S. & Ouzounis, C. A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584 (2002)
Rawlings, N. D., Morton, F. R. & Barrett, A. J. MEROPS: the peptidase database. Nucleic Acids Res. 34, D270–D272 (2006)
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994)
Kumar, S., Tamura, K. & Nei, M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5, 150–163 (2004)
Acknowledgements
This investigation was mainly supported by the Chinese National High-Tech Program (863 Program) (2004AA2Z1010, 2006AA02Z335, 2006AA02Z318, 2007AA02Z153), the Chinese National Key Project for Basic Research (973 Project) (2006CB708510, 2007CB513100), the Chinese Academy of Sciences, the Shanghai Municipal Commission for Science and Technology (04DZ14010, 055407031, 06JC14059, 07QA14043, 07DZ22915), and the National Natural Science Foundation of China. Support from the US National Institute of Allergy and Infectious Diseases (award number AI39461), the National Science and Engineering Research Council of Canada (OGP0046506), a International Collaborative Research Grants award from the National Health and Medical Research Council of Australia, and the Wellcome Trust, UK, is also gratefully acknowledged. The Shanghai Supercomputer Center kindly provided computational facilities for some of the data analysis. The authors wish to thank P. De Jong and his colleagues for the BAC libraries construction of S. japonicum and N. M. El-Sayed for his contribution on the collaboration between the S. japonicum and S. mansoni sequencing consortia.
Author Contributions Y. Zhou, H.Z., F.L., W. Hu, Z.-Q.W., G.L. and S.R. contributed equally to this work.
Author information
Authors and Affiliations
Consortia
Corresponding authors
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary References, Supplementary Tables 1-31 and Supplementary Figures 1-16. (PDF 7832 kb)
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution-Non-Commercial-Share Alike licence (http://creativecommons.org/licenses/by-nc-sa/3.0/), which permits distribution, and reproduction in any medium, provided the original author and source are credited. This license does not permit commercial exploitation, and derivative works must be licensed under the same or similar licence.
About this article
Cite this article
The Schistosoma japonicum Genome Sequencing and Functional Analysis Consortium. The Schistosoma japonicum genome reveals features of host–parasite interplay. Nature 460, 345–351 (2009). https://doi.org/10.1038/nature08140
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature08140
This article is cited by
-
Comprehensive analysis of miRNA profiling in Schistosoma mekongi across life cycle stages
Scientific Reports (2024)
-
Evolution of tetraspanin antigens in the zoonotic Asian blood fluke Schistosoma japonicum
Parasites & Vectors (2023)
-
Construction and characterization of microsatellite markers for the Schistosoma japonicum isolate from a hilly area of China based on whole genome sequencing
Parasitology Research (2023)
-
Analysis of rhodopsin G protein-coupled receptor orthologs reveals semiochemical peptides for parasite (Schistosoma mansoni) and host (Biomphalaria glabrata) interplay
Scientific Reports (2022)
-
Serum proteomic profiling in patients with advanced Schistosoma japonicum-induced hepatic fibrosis
Parasites & Vectors (2021)